ARTÍCULO
First genome size assessments in Carduncellus and its related genera Femeniasia and Phonus (Asteraceae, Cardueae), with data on 21 taxa
TERESA GARNATJE1, ORIANE HIDALGO1, JOAN VALLÈS2, SÒNIA GARCIA1, ÀNGEL ROMO1 & ROSER VILATERSANA1
1 Institut Botànic de Barcelona (IBB, CSIC-Ajuntament de Barcelona), pg. del Migdia, s/n, Parc de Montjuïc, ES-08038 Barcelona, Catalonia, Spain
2 Laboratori de Botànica - Unitat associada CSIC, Facultat de Farmàcia i Ciències de l’Alimentació - Institut de la Biodiversitat IRBio, Universitat de Barcelona, av. Joan XXIII, 27-31, 08028 Barcelona
ORCID iD. T. GARNATJE: https://orcid.org/0000-0001-6295-6217, O. HIDALGO: https://orcid.org/0000-0002-1547-8627, J. VALLÈS: https://orcid.org/0000-0002-1309-3942, S. GARCIA: http://orcid.org/0000-0002-3143-0527, À. ROMO: https://orcid.org/0000-0001-8135-8570, R. VILATERSANA: https://orcid.org/0000-0002-5106-8764
Author for correspondence: T. Garnatje (tgarnatje@ibb.csic.es)
Editor: Jordi López-Pujol
ABSTRACT
First genome size assessments in Carduncellus and its related genera Femeniasia and Phonus (Asteraceae, Cardueae), with data on 21 taxa.— Genome size of 18 species of the genus Carduncellus, two species of the related genus Phonus and the monotypic genus Femeniasia (F. balearica) has been assessed by flow cytometry for the first time. Ploidy levels were assigned using genome size data together with previously reported chromosome counts. A phylogenetic framework was built to visualize how cytogenetic traits distributed across taxa. The results confirmed three ploidy levels (2x, 4x and 6x), with a predominance of diploids. The 2C values ranged from 3.24 pg in Carduncellus calvus to 11.16 pg in C. eriocephalus, whereas monoploid genome size (1Cx) ranged from 1.29 pg in C. duvauxii (4x) to 2.30 pg in Phonus rhiphaeus (2x). The mean 1Cx for tetraploids was lower than for diploids. For each ploidy level, genome size values of Carduncellus, Femeniasia and Phonus were found to be higher than those of Carthamus. This result is consistent with a trend frequently observed in plants, of higher genome sizes in long life cycle taxa compared to short-lived relatives.
KEY WORDS: 2C-values; Carthamus-Carduncellus complex; DNA amount; life-cycle; polyploidy; ploidy level.
RESUMEN
Primeras medidas del tamaño del genoma en Carduncellusy los géneros afines Femeniasia y Phonus (Asteraceae, Cardueae), con datos para 21 táxones.— El tamaño del genoma de 18 especies del género Carduncellus, dos especies de los géneros relacionados, Phonus y el género monotípico Femeniasia (F. balearica) ha sido medido por primera vez mediante citometría de flujo. Los niveles de ploidía se asignaron utilizando datos de tamaño del genoma junto con los recuentos de cromosomas previamente reportados. Se construyó un marco filogenético para visualizar la distribución de las características citogenéticas de los táxones. Los resultados confirmaron tres niveles de ploidía (2x, 4x y 6x), con un predominio de los táxones diploides. Los valores de 2C oscilaron entre 3,24 pg en Carduncellus calvus y 11,16 pg en C. eriocephalus, mientras que el tamaño del genoma monoploide (1Cx) osciló entre 1,29 pg en C. duvauxii (4x) y 2,30 pg en Phonus rhiphaeus (2x). La media de los valores 1Cx para los tetraploides fue menor que para los diploides. Los valores de tamaño del genoma de Carduncellus, Femeniasia y Phonus fueron más elevados que los de Carthamus dentro del mismo nivel de ploidía. Este resultado concuerda con una tendencia frecuentemente observada en plantas en la que los táxones con ciclos de vida largos presentan tamaños del genoma más elevados que los táxones relacionados que poseen ciclos de vida cortos.
PALABRAS CLAVE: cantidad de ADN; ciclo vital; complejo Carthamus-Carduncellus; nivel de ploidía; poliploidía; valores 2C.
Received 9 October 2020; accepted 26 January 2021; published on line 18 June 2021
Cómo citar este artículo / Citation: Garnatje, T., Hidalgo, O., Vallès, J., Garcia, S., Romo, A. & Vilatersana, R. 2021. First genome size assessments in Carduncellus and its related genera Femeniasia and Phonus (Asteraceae, Cardueae), with data on 21 taxa. Collectanea Botanica 40: e004. https://doi.org/10.3989/collectbot.2021.v40.004
Copyright: © 2021 CSIC. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License.
CONTENIDOS
INTRODUCTIONTop
Genome size has been revealed as a powerful tool for studying allopolyploidy and hybridization in the genus Carthamus L. (Garnatje et al., 2006Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050) and evolutionary processes in the Asteraceae as a whole (e.g. Torrell & Vallès, 2001Torrell, M. & Vallès, J. 2001. Genome size in 21 Artemisia L. species (Asteraceae, Anthemideae): Systematic, evolutionary, and ecological implications. Genome 44: 231–238. https://doi.org/10.1139/g01-004; Bureš et al., 2004Bureš, P., Wang, Y.-F., Horová, L. & Suda, J. 2004. Genome size variation in central European species of Cirsium (Compositae) and their natural hybrids. Annals of Botany 94: 353–363. https://doi.org/10.1093/aob/mch151; Bancheva & Greilhuber, 2006Bancheva, S. & Greilhuber, J. 2006. Genome size in Bulgarian Centaurea s.l. (Asteraceae). Plant Systematics and Evolution 257: 95–117. https://doi.org/10.1007/s00606-005-0384-7; Garcia et al., 2006Garcia, S., Garnatje, T., Twibell, J. D. & Vallès, J. 2006. Genome size variation in the Artemisia arborescens complex (Asteraceae, Anthemideae) and its cultivars. Genome 49: 244–253. https://doi.org/10.1139/g05-105, 2008Garcia, S., Canela M. Á., Garnatje, T., McArthur, E. D., Pellicer, J., Sanderson, S. C. & Vallès, J. 2008. Evolutionary and ecological implications of genome size in the North American endemic sagebrushes and allies (Artemisia, Asteraceae). Biological Journal of the Linnean Society 94: 631–649. https://doi.org/10.1111/j.1095-8312.2008.01001.x; Suda et al., 2007Suda, J., Krahulcová, A., Trávnícek, P., Rosenbaumová, R., Peckert, T. & Krahulec, F. 2007. Genome size variation and species relationships in Hieracium sub-genus Pilosella (Asteraceae) as inferred by flow cytometry. Annals of Botany 100: 1323–1335. https://doi.org/10.1093/aob/mcm218; Hidalgo et al., 2008Hidalgo, O., Garcia-Jacas, N., Garnatje, T., Romashchenko, K., Susanna, A. & Siljak-Yakovlev, S. 2008. Extreme environmental conditions and phylogenetic inheritance: systematics of Myopordon and Oligochaeta (Asteraceae, Cardueae-Centaureinae). Taxon 57: 769–778. https://doi.org/10.1002/tax.573009, 2017Hidalgo, O., Vitales, D., Vallès, J., Garnatje, T., Siljak-Yakovlev, S., Leitch, I. J. & Pellicer J. 2017. Cytogenetic insights into an oceanic island radiation: The dramatic evolution of pre-existing traits in Cheirolophus (Asteraceae: Cardueae: Centaureinae). Taxon 66: 146–157. https://doi.org/10.12705/661.8; Pellicer et al., 2010Pellicer, J., Garcia, S., Canela, M. Á., Garnatje, T., Korobkov, A. A., Twibell, J. D. & Vallès, J. 2010. Genome size dynamics in Artemisia L. (Asteraceae): following the track of polyploidy. Plant Biology 12: 820–830. https://doi.org/10.1111/j.1438-8677.2009.00268.x; Trávníček et al., 2013Trávníček, P., Kirschner, J., Chudáčková, H., Rooks, F. & Štěpánek, J. 2013. Substantial genome size variation in Taraxacum stenocephalum (Asteraceae, Lactuceae). Folia Geobotanica 48: 271–284. https://doi.org/10.1007/s12224-013-9151-7; Pegoraro et al., 2020Pegoraro, L., Baker, E. C., Aeschimann, D., Balant, M., Douzet, R., Garnatje, T., Guignard, M. S., Leitch, I. J., Leitch, A. R., Palazzesi, L., Theurillat, J. P., Hidalgo, O. & Pellicer, J. 2020. The correlation of phylogeny, elevation and ploidy on the incidence of apomixis in Asteraceae of the European Alps. Botanical Journal of the Linnean Society 194: 410–422. https://doi.org/10.1093/botlinnean/boaa058; Vitales et al., 2020Vitales, D., Álvarez, I., Garcia, S., Hidalgo, O., Nieto Feliner, G., Pellicer, J., Vallès, J. & Garnatje, T. 2020. Genome size variation at constant chromosome number is not correlated with repetitive DNA dynamism in Anacyclus (Asteraceae). Annals of Botany 125: 611–623. https://doi.org/10.1093/aob/mcz183).
Together with Carthamus, the genera Carduncellus Adans., Femeniasia Susanna and Phonus Hill constitute the Carthamus-Carduncellus complex (Vilatersana et al., 2000aVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000a. Generic delimitation and phylogeny of the Carduncellus–Carthamus complex (Asteraceae) based on ITS sequences. Plant Systematics and Evolution 221: 89–105. https://doi.org/10.1007/BF01086383; Vilatersana, 2002Vilatersana, R. 2002. Delimitació genèrica del complex Carthamus-Carduncellus: un assaig de biosistemàtica i sistemàtica molecular. PhD Thesis, Universitat de Barcelona, Barcelona.). Carthamus comprises 18 annual species growing in disturbed habitats of the eastern part of Mediterranean basin and western Asia. Sister to the genus Carthamus, the clade comprising the genera Carduncellus (ca. 26 species, from North Africa and the Iberian Peninsula), Femeniasia (one species from Menorca, Balearic Islands) and Phonus (two species from North Africa and Iberian Peninsula) is made of perennial species which grow in few disturbed habitats (Vilatersana et al., 2000aVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000a. Generic delimitation and phylogeny of the Carduncellus–Carthamus complex (Asteraceae) based on ITS sequences. Plant Systematics and Evolution 221: 89–105. https://doi.org/10.1007/BF01086383; López González, 2012López González, G. 2012. Sobre la clasificación del complejo Carthamus-Carduncellus (Asteraceae, Cardueae-Centaureinae) y su tratamiento en Flora iberica. Acta Botanica Malacitana 37: 79–92. https://doi.org/10.24310/abm.v37i0.2669). However, generic circumscription in the Carthamus-Carduncellus complex is still a matter of debate. In the last two decades, different taxonomic treatments resulted in the recognition of (i) the four genera Carthamus, Carduncellus, Femeniasia and Phonus, the treatment we followed in this study (Vilatersana et al., 2000aVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000a. Generic delimitation and phylogeny of the Carduncellus–Carthamus complex (Asteraceae) based on ITS sequences. Plant Systematics and Evolution 221: 89–105. https://doi.org/10.1007/BF01086383), (ii) an expanded genus Carthamus encompassing the three other genera (Greuter, 2003Greuter, W. 2003. The Euro+Med treatment of Cardueae (Compositae) – generic concepts and required new names. Willdenowia 33: 49–61. https://doi.org/10.3372/wi.33.33104), and (iii) two genera, Carthamus and Carduncellus, the delimitation of the latter being extended to include Femeniasia and Phonus (López González, 2012López González, G. 2012. Sobre la clasificación del complejo Carthamus-Carduncellus (Asteraceae, Cardueae-Centaureinae) y su tratamiento en Flora iberica. Acta Botanica Malacitana 37: 79–92. https://doi.org/10.24310/abm.v37i0.2669).
In addition to their distinct life cycles and habitat preference, the genera of the Carthamus-Carduncellus complex also have different karyological and cytogenetic profiles (Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x). The evolution of Carthamus involved descending dysploidy (x = 12, 11, 10) and polyploidy (2x, 4x, 6x; Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x; Garnatje et al., 2006Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050). This genus presents 2C values from 2.26 to 7.46 pg and monoploid genome sizes (1Cx) from 1.13 to 1.53 pg (Garnatje et al., 2006Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050). Genome size of allopolyploids was found to be the sum of their parental species, or slightly inferior (Garnatje et al., 2006Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050). The clade constituted by Carduncellus, Femeniasia and Phonus presents a constant base chromosome number of x = 12, with 2x cytotypes for Femeniasia and Phonus, and 2x to 6x cytotypes for Carduncellus (Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x; Vilatersana, 2002Vilatersana, R. 2002. Delimitació genèrica del complex Carthamus-Carduncellus: un assaig de biosistemàtica i sistemàtica molecular. PhD Thesis, Universitat de Barcelona, Barcelona.). In Carduncellus, diploids predominate especially among the endemic species occupying narrow areas, tetraploids are relatively frequent, while triploids and hexaploids are found more sporadically (López González, 1990López González, G. 1990. Acerca de la clasificación natural del género Carthamus L., s.l. Anales del Jardín Botánico de Madrid 47: 11–34. ; Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x; Vilatersana, 2002Vilatersana, R. 2002. Delimitació genèrica del complex Carthamus-Carduncellus: un assaig de biosistemàtica i sistemàtica molecular. PhD Thesis, Universitat de Barcelona, Barcelona.). B chromosomes are not rare in these species, indicating high genome dynamism (Vilatersana, 2002Vilatersana, R. 2002. Delimitació genèrica del complex Carthamus-Carduncellus: un assaig de biosistemàtica i sistemàtica molecular. PhD Thesis, Universitat de Barcelona, Barcelona.). No data on genome size is available so far for any taxa of the Carduncellus-Femeniasia-Phonus clade.
This study aims at improving our understanding of cytogenetic evolution within the Carthamus-Carduncellus complex. We provide the first genome size data for the genera Carduncellus, Femeniasia and Phonus and discuss cytotype diversity in the light of phylogenetic and ecological contexts.
MATERIALS AND METHODSTop
Plant material
The sampling comprises a total of 41 populations of 18 species of genus Carduncellus, including three subspecies, two species of Phonus and one population of Femeniasia balearica (J. J. Rodr.) Susanna. Origin, collectors and dates are shown in Table 1. Voucher specimens for each population are deposited in the herbarium BC (Botanical Institute of Barcelona).
| Table 1. Origin, collectors and dates of the studied material. Vouchers are deposited in the herbarium BC. |
|
Taxon (code)
|
Origin, collectors and date
|
|
Carduncellus caeruleus (L.) C. Presl. (1)
|
Spain, Málaga: Tolox, Garcia-Jacas, Susanna 1610 & Vilatersana, 22.VI.1996
|
|
Carduncellus caeruleus (2)
|
Morocco, Fes: Oued Zloul valley near Ahermoumou, Garnatje, Susanna 1801 & Vilatersana, 18.VI.1997
|
|
Carduncellus caeruleus (3)
|
Spain, Córdoba: between Jauja and Puente Genil, Vilatersana 59, 8.IV.1998
|
|
Carduncellus calvus Boiss. & Reut.
|
Morocco, Tahanaout, El Fellah, Romo 14025 & Vilatersana, 17.VI.2006
|
|
Carduncellus catrouxii Emb.
|
Morocco, Mgoun area: Ouzighimt-Tal, Finckh & Staudinger 859, 1.VII.2002 (Herbarium Hamburgense)
|
|
Carduncellus cuatrecasasii G. López (1)
|
Spain, Jaén: Sierra del Ahillo, Sanz & Vilatersana 506, 12.VI.2005
|
|
Carduncellus cuatrecasasii (2)
|
Spain, Jaén: Sierra de Segura, Sanz & Vilatersana 527, 18.VI.2005
|
|
Carduncellus dianius Webb. (1)
|
Spain, Valencia: Xàbia, Cap de Sant Antoni, Garcia-Jacas, Susanna 1479 & Vilatersana, 17.VI.1995
|
|
Carduncellus dianius (2)
|
Spain, Balearic Islands, Eivissa: Cala Ximena, Garnatje & Vilatersana 402, 15.IV.2004
|
|
Carduncellus duvauxii Batt. et Trab.
|
Morocco, Ujdah: Bouarfa, Garnatje, Susanna 1779 & Vilatersana, 16.VI.1997
|
|
Carduncellus eriocephalus Boiss.
|
Morocco, Ujdah: Bouanane, Garnatje, Susanna 1785 & Vilatersana, 16.VI.1997
|
|
Carduncellus fruticosus (Maire) Hanelt
|
Morocco, Ouarzazate: River Todrha, Benedí, G. Montserrat & J. M. Montserrat 2407, 5.VI.1980
|
|
Carduncellus hispanicus Boiss. ex DC. subsp. araneosus (Boiss. & Reut.) G. López
|
Spain, Toledo: between Huertas de Valdecábanos and Cabañas de Yepes, Sanz & Vilatersana 529, 18.VI.2005
|
|
Carduncellus hispanicus subsp. hispanicus
|
Spain, Almería: Sierra de Gádor, Sanz & Vilatersana 486, 8.VI.2005
|
|
Carduncellus hispanicus subsp. intercedens (Degen & Hervier) G. López (1)
|
Spain, Alacant: Serra de Bèrnia, Garnatje & Vilatersana 450, 22.VI.2005
|
|
Carduncellus hispanicus subsp. intercedens (2)
|
Spain, Murcia: Sierra de la Muela, Garnatje & Vilatersana 460, 24.VI.2005
|
|
Carduncellus hispanicus subsp. intercedens (3)
|
Spain, Granada: Sierra de Baza, Sanz & Vilatersana 516, 15.VII.2005
|
|
Carduncellus lucens Ball (1)
|
Morocco, Oikaimeden, Romo 13929 & Vilatersana, 13.VI.2006
|
|
Carduncellus lucens (2)
|
Morocco, Oikaimeden, Romo 13930 & Vilatersana, 13.VI.2006
|
|
Carduncellus lucens (3)
|
Morocco: Oukaimeden, Romo 13927 & Vilatersana, 12.VI.2006
|
|
Carduncellus mareoticus (Del.) Hanelt (1)
|
Egypt, Alexandria: road Alexandria-Marsah Matruh, Susanna 1860 & Vilatersana, 22.VI.1996
|
|
Carduncellus mareoticus (2)
|
Egypt, Alexandria: El Amiriya, Susanna 1850 & Vilatersana, 7.VI.1998
|
|
Carduncellus mareoticus (3)
|
Egypt, Alexandria: New Bourg-el-Arab, Susanna 1846 & Vilatersana, 7.VI.1998
|
|
Carduncellus mitissimus DC.
|
Spain, Navarra: between Burgui and Navascués, Carretero & Vilatersana 72, 7.VII.2000
|
|
Carduncellus monspelliensium All. (1)
|
Spain, Tarragona: Serra del Monsant, Vilatersana 18, 23.IX.1995.
|
|
Carduncellus monspelliensium (2)
|
Spain, Soria: Montejo de Tiermes, Garcia-Jacas & Susanna 2223, 27.VII.2001
|
|
Carduncellus monspelliensium (3)
|
Spain, Tarragona: Morera del Montsant, Vilatersana 17, 18.X.1997
|
|
Carduncellus monspelliensium (4)
|
Spain, Tarragona: Santa Coloma de Queralt, Garcia-Jacas, Susanna & Vilatersana 10, 10.VI.1995
|
|
Carduncellus monspelliensium (5)
|
Spain, Tarragona: Sant Magí de Brufaganya, Del Rey & Vilatersana 702, 8.VIII.2006
|
|
Carduncellus pectinatus DC.
|
Morocco: Azerzou, between Tanourdi and Ait-Mouli, Romo 14028 & Vilatersana, 17.VI.2006
|
|
Carduncellus pinnatus (Desf.) DC.
|
Morocco: Ouarzazate, Tizi n’Tichka Pass, 2169 m, Romo 13910 & Vilatersana, 12.VI.2006
|
|
Carduncellus pomelianus Batt. (1)
|
Morocco, Middle Atlas: near Djebel Amjoud, J. Molero, J. M. Montserrat 6787& L. Sáez, 24.VII.2000
|
|
Carduncellus pomelianus (2)
|
Morocco: Boumia, Romo 14060 & Vilatersana, 18.VI.2006
|
|
Carduncellus reboudianus Batt. (1)
|
Morocco, Ksar es Souk: Tizi n’Talrhem, Garnatje, Susanna 1788 & Vilatersana, 17.VI.1997
|
|
Carduncellus reboudianus (2)
|
Morocco: between Ait Toughach and Zaida, Romo 14031 & Vilatersana, 17.VI.2006
|
|
Carduncellus rhaponticoides Coss. & Dur. (1)
|
Morocco, Oukaïmeden: Col du Tizrag, G. López 8958 & F. Muñoz Garmendia, 11.VII.1984
|
|
Carduncellus rhaponticoides (2)
|
Morocco: between Oualeger and the Zad pass, Romo 14054 & Vilatersana, 17.VI.2006
|
|
Femeniasia balearica (J. J. Rodr.) Susanna
|
Spain, Balearic Islands, Menorca: Mongofre Vell, J. M. Montserrat 2802, 5.VII.1991
|
|
Phonus arborescens (L.) G. López (1)
|
Spain, Almería: Sierra de Gádor near Félix, J. M. Montserrat, 27.VII.1990
|
|
Phonus arborescens (2)
|
Spain, Almería: between Roquetas and Félix, Garcia-Jacas, Susanna 1611 & Vilatersana, 24.VI.1996
|
|
Phonus rhiphaeus (Font Quer & Pau) G. López
|
Morocco, Al Hoceima: Tleta Oued Laou between Tarerha and Azenti, J. M. Montserrat 4360, Pallàs & Veny, 23.VI.1993
|
|
DNA content assessment
Fresh young leaves of the plants studied were co-chopped using a razor blade with an internal standard in the proportions 2:1 in 1200 μl of LB01 buffer (Doležel et al., 1989Doležel, J., Binarová, P. & Lucretti, S. 1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia Plantarum 3: 113–120. https://doi.org/10.1007/BF02907241) with 0.5% of Triton X-100 and supplemented with 100 µg/ml ribonuclease A (RNase A, Boehringer, Meylan, France) in a plastic Petri dish. Pisum sativum L. ‘Express Long’ (2C = 8.37 pg) and Petunia hybrida Vilm. ‘PxPc6’ (2C = 2.85 pg) were used as internal standards and were first analysed separately in 600 μl of LB01 buffer to locate their peak positions. Nuclei were filtered through a 70-μm nylon filter in order to eliminate cell debris before the addition of 36 μl of propidium iodide (1 mg/ml, solution in water; Invitrogen Eugene, Oregon, USA). Samples were kept on ice before measurement. For each population (Table 1), two samples of each individual were prepared and measured independently. Fluorescence analysis was carried out using an Epics XL flow cytometer (Coulter Corporation, Hialeah, Florida, USA) at the Centres Científics i Tecnològics de la Universitat de Barcelona with the standard configuration as described in Garnatje et al. (2006Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050). Acquisition was stopped at 8000 nuclei. The DNA content was calculated for 10 of the aforementioned runs, assuming a linear correlation between the fluorescence signals (of the stained nuclei) and DNA amount. Mean and standard deviations were calculated for 2C values of each population based on five individuals.
Phylogenetic framework
The nuclear ribosomal dataset includes ITS1 and ITS2 regions for 23 species, including two outgroups. ITS sequences of 17 species were available from GenBank and four taxa were sequenced for this study following the same protocol as described in Barres et al. (2013Barres, L., Sanmartín, I., Anderson, C. L., Susanna, A., Buerki, S., Galbany-Casals, M. & Vilatersana, R. 2013. Reconstructing the evolution and biogeographic history of tribe Cardueae (Compositae). American Journal of Botany 100: 867–882. https://doi.org/10.3732/ajb.1200058). Carthamus glaucus M. Bieb. and C. oxyacantha M. Bieb. were chosen as outgroup based on published phylogeny by Vilatersana et al. (2000aVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000a. Generic delimitation and phylogeny of the Carduncellus–Carthamus complex (Asteraceae) based on ITS sequences. Plant Systematics and Evolution 221: 89–105. https://doi.org/10.1007/BF01086383). DNA sequences were edited with Chromas v2.6.4 (Technelysium PTy, Tewantin, Queensland, Australia) and Bioedit v7.0.9 (Hall, 1999Hall, T. A. 1999. BioEdit, A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.) and aligned visually. The aligned matrix is available from the corresponding author. The General Time Reversible model (GTR + G) was chosen for ITS nrDNA dataset based on AIC criterion implemented in jModeltest v2.1.2 (Darriba et al., 2012Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. https://doi.org/10.1038/nmeth.2109). Markov Chain Monte Carlo (MCMC) analysis was carried out in MrBayes v3.2.6 (Ronquist et al., 2012Ronquist, F., Teslenko, M., Mark, P., Ayres, D., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A. & Huelsenbeck, J. P. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029) for 2,000,000 generation sampling every 100 generations. The first 25% of the trees were discarded as the ‘burn-in’ period, after confirming that the average standard deviation of the split frequencies was <0.01, and the potential scale reduction factor approached 1.0 for all parameters. The remaining samples were pooled to construct a 50% majority rule consensus tree. The resulting summary trees were visualised in Figtree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree).
Bar plots showing the distribution of genome size values across the taxa of the phylogenetic inference were generated with the package phytools (Revell, 2012Revell, L. J. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. https://doi.org/10.1111/j.2041-210X.2011.00169.x; implemented in R v3.2.2; R Core Team, 2016R Core Team 2016. R: a language and environment for statistical computing. Foundation for Statistical Computing, Vienna. Version 3.6.2. Retrieved December 12, 2019, from http://www.R-project.org). A graph illustrating the distribution of mean genome size values for the species of the Carthamus-Carduncellus complex at different ploidy levels was generated with the package ggplot2 (Wickham, 2016Wickham, H. 2016. ggplot2: elegant graphics for data analysis. Springer, New York. https://doi.org/10.1007/978-3-319-24277-4; implemented in R) using the new genome size assessments together with the previously published data of Garnatje et al. (2006Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050).
Statistical analyses
One-way ANOVA was carried out in order to test the 1Cx differences between ploidy levels and between Carthamus and Carduncellus lineages. The analyses were performed with XLSTAT 2018.7 (Addinsoft Inc.).
RESULTS AND DISCUSSIONTop
Nuclear DNA amount (2C in pg and 1C in Mbp), chromosome numbers counted for the same populations (Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x), ploidy levels and 1Cx values (pg) are shown in Table 2 and Fig. 1. The GenBank accession numbers for new sequences are included in Table 3. To our knowledge and according to recently updated Asteraceae and plant genome size databases (respectively https://www.asteraceaegenomesize.com, Vitales et al., 2019Vitales, D., Fernández, P., Garnatje, T. & Garcia, S. 2019. Progress in the study of genome size evolution in Asteraceae: analysis of the last update. Database 2019: baz098. https://doi.org/10.1093/database/baz098; and https://cvalues.science.kew.org, Pellicer & Leitch, 2020Pellicer, J. & Leitch, I. J. 2020. The Plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytologist 226: 301–305. https://doi.org/10.1111/nph.16261, both accessed August 11, 2020), these are the first nuclear DNA amount assessments for the 21 species and three genera studied.
| Table 2. Nuclear DNA content and other karyological characteristics of the studied species. |
|
Taxon
|
2C ± SD
(pg) 1
|
1C (Mbp) 2
|
1Cx (pg) 3
|
2 n 4
|
x 5
|
Standard
|
|
Carduncellus caeruleus (1)
|
6.48 ± 0.04
|
3169
|
1.62
|
48
|
4
|
Petunia
|
|
Carduncellus caeruleus (2)
|
6.29 ± 0.04
|
3076
|
1.57
|
48
|
4
|
Petunia
|
|
Carduncellus caeruleus (3)
|
6.32 ± 0.16
|
3091
|
1.58
|
-
|
4*
|
Petunia
|
|
Carduncellus calvus
|
3.24 ± 0.21
|
1584
|
1.62
|
-
|
2*
|
Petunia
|
|
Carduncellus catrouxii
|
7.07 ± 0.09
|
3457
|
1.77
|
-
|
4*
|
Petunia
|
|
Carduncellus cuatrecasasii (1)
|
4.24 ± 0.15
|
2073
|
2.12
|
-
|
2*
|
Petunia
|
|
Carduncellus cuatrecasasii (2)
|
3.32 ± 0.03
|
1624
|
1.66
|
-
|
2*
|
Petunia
|
|
Carduncellus dianius (1)
|
3.45 ± 0.01
|
1687
|
1.73
|
24
|
2
|
Petunia
|
|
Carduncellus dianius (2)
|
3.43 ± 0.02
|
1677
|
1.72
|
-
|
2*
|
Pisum
|
|
Carduncellus duvauxii
|
5.15*
|
2518
|
1.29
|
48
|
4
|
Petunia
|
|
Carduncellus eriocephalus
|
11.16 ± 0.41
|
5457
|
1.86
|
72
|
6
|
Pisum
|
|
Carduncellus fruticosus
|
4.28 ± 0.05
|
2093
|
2.14
|
24
|
2
|
Petunia
|
|
Carduncellus hispanicus subsp. araneosus
|
3.50 ± 0.13
|
1712
|
1.75
|
24
|
2
|
Petunia
|
|
Carduncellus hispanicus subsp. hispanicus
|
4.33 ± 0.30
|
2177
|
2.17
|
24
|
2
|
Petunia
|
|
Carduncellus hispanicus subsp. intercedens (1)
|
7.00 ± 0.05
|
3423
|
1.75
|
-
|
4*
|
Pisum
|
|
Carduncellus hispanicus subsp. intercedens (2)
|
7.31 ± 0.29
|
3575
|
1.83
|
-
|
4*
|
Pisum
|
|
Carduncellus hispanicus subsp. intercedens (3)
|
7.09 ± 0.23
|
3467
|
1.77
|
-
|
4*
|
Petunia
|
|
Carduncellus lucens (1)
|
3.56 ± 0.09
|
1741
|
1.78
|
-
|
2*
|
Pisum
|
|
Carduncellus lucens (2)
|
3.27 ± 0.19
|
1599
|
1.64
|
-
|
2*
|
Pisum
|
|
Carduncellus lucens (3)
|
3.27*
|
1599
|
1.64
|
-
|
2*
|
Petunia
|
|
Carduncellus mareoticus (1)
|
4.40 ± 0.06
|
2152
|
2.20
|
-
|
2*
|
Petunia
|
|
Carduncellus mareoticus (2)
|
4.56*
|
2230
|
2.28
|
24
|
2
|
Pisum
|
|
Carduncellus mareoticus (3)
|
4.52 ± 0.09
|
2210
|
2.26
|
-
|
2*
|
Petunia
|
|
Carduncellus mitissimus
|
3.59 ± 0.02
|
1756
|
1.80
|
-
|
2*
|
Petunia
|
|
Carduncellus monspelliensium (1)
|
7.38*
|
3609
|
1.85
|
48
|
4
|
Petunia
|
|
Carduncellus monspelliensium (2)
|
6.94 ± 0.11
|
3394
|
1.74
|
-
|
4*
|
Petunia
|
|
Carduncellus monspelliensium (3)
|
7.14*
|
3492
|
1.79
|
48
|
4
|
Petunia
|
|
Carduncellus monspelliensium (4)
|
7.22 ± 0.13
|
3531
|
1.81
|
-
|
4*
|
Petunia
|
|
Carduncellus monspelliensium (5)
|
7.74 ± 0.77
|
3638
|
1.94
|
-
|
4*
|
Petunia
|
|
Carduncellus pectinatus
|
3.43 ± 0.18
|
1677
|
1.72
|
-
|
2*
|
Pisum
|
|
Carduncellus pinnatus
|
4.22 ± 0.56
|
2064
|
2.11
|
-
|
2*
|
Petunia
|
|
Carduncellus pomelianus (1)
|
3.41 ± 0.06
|
1668
|
1.71
|
-
|
2*
|
Petunia
|
|
Carduncellus pomelianus (2)
|
3.54 ± 0.16
|
1731
|
1.77
|
-
|
2*
|
Pisum
|
|
Carduncellus reboudianus (1)
|
6.55 ± 0.12
|
3203
|
1.64
|
48
|
4
|
Petunia
|
|
Carduncellus reboudianus (2)
|
6.71 ± 0.16
|
3281
|
1.68
|
-
|
4*
|
Pisum
|
|
Carduncellus rhaponticoides (1)
|
6.79 ± 0.12
|
3320
|
1.70
|
-
|
4*
|
Petunia
|
|
Carduncellus rhaponticoides (2)
|
6.96 ± 0.18
|
3403
|
1.74
|
-
|
4*
|
Pisum
|
|
Femeniasia balearica
|
3.84 ± 0.03
|
1878
|
1.92
|
24
|
2
|
Petunia
|
|
Phonus arborescens (1)
|
4.54 ± 0.07
|
2220
|
2.27
|
24
|
2
|
Petunia
|
|
Phonus arborescens (2)
|
4.56 ± 0.04
|
2230
|
2.28
|
-
|
2*
|
Petunia
|
|
Phonus rhiphaeus
|
4.60 ± 0.06
|
2249
|
2.30
|
24
|
2
|
Petunia
|
1 Holoploid genome size (2C) values in pg with standard deviation. Asterisk indicates when a single measurement has been done.
2 Holoploid genome size (1C) values in Mbp. 1 pg = 978 Mbp (Doležel et al., 2003Doležel, J., Bartos, J., Voglmayr, H. & Greilhuber, J. 2003. Nuclear DNA content and genome size of trout and human. Cytometry 51A: 127–128. https://doi.org/10.1002/cyto.a.10013).
3 Monoploid genome size (1Cx) values in pg.
4 Chromosome counts from Vilatersana et al. (2000b) corresponding to the same accessions measured for genome size.
5 Ploidy levels. Asterisk indicates when chromosome counts from other accessions than the one measured for genome size were used to infer the ploidy level.
|
|
|
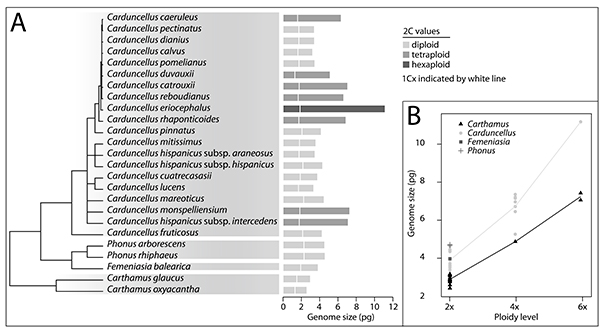
Figure 1. Distribution of genome size and ploidy levels across species of the Carthamus-Carduncellus complex: (A), molecular phylogeny of Carduncellus and related genera Femeniasia and Phonus. Bars represent mean 2C value per species, with 1Cx indicated by a white line. Different colour intensities of bars depict ploidy levels; (B), distribution of mean genome size 2C values for species of the Carthamus-Carduncellus complex at diploid, tetraploid and hexaploid levels. Lines connect the mean values per genus and ploidy level. Genome size values for Carthamus were obtained from Garnatje et al. (2006Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050).
[View full size] [Descargar tamaño completo] |
|
| Table 3. GenBank accession numbers for the species sequenced in this study. |
|
Taxon
|
ITS1
|
ITS2
|
|
Carduncellus catrouxii
|
MW209004
|
MW208954
|
|
Carduncellus lucens
|
MW209005
|
MW208955
|
|
Carduncellus pectinatus
|
MW209007
|
MW208957
|
|
Carduncellus reboudianus
|
MW209006
|
MW208956
|
|
Genome size and ploidy level diversity in the Carduncellus-Femeniasia-Phonus clade
Genome size values (2C) range from 3.24 pg in Carduncellus calvus Boiss. & Reut., a diploid species endemic to Maghreb region (Morocco and Algeria), to 11.16 pg in a hexaploid accession of C. eriocephalus Boiss., a species showing several ploidy levels (Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x). This species occurs in the Maghreb region, reaching Egypt. Femeniasia balearica, endemic to Menorca (Balearic Islands), presents a 2C value of 3.84 pg, and Phonus, 2C values from 4.54 to 4.60 pg. Monoploid genome size (1Cx) ranges from 1.29 pg in a tetraploid accession of C. duvauxii Batt. et Trab. (2n = 48; Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x), which has also been reported as a diploid by López González (1990López González, G. 1990. Acerca de la clasificación natural del género Carthamus L., s.l. Anales del Jardín Botánico de Madrid 47: 11–34. ), to 2.28 in C. mareoticus (Del.) Hanelt, a diploid species endemic from the northern part of Egypt and Libya. Femeniasia balearica displays a 1Cx value of 1.92 and the 1Cx values oscillate between 2.27 and 2.30 in the two studied species of the genus Phonus, all of them being diploid.
Three ploidy levels have been found (2x, 4x and 6x) in genus Carduncellus, some of which have been inferred from genome size values. Fifteen taxa are diploid, seven tetraploid and only one (C. eriocephalus) is a hexaploid. It is to note that a triploid chromosome count was also reported in this same Moroccan accession of C. eriocephalus (Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x), indicating within-population cytotype diversity. In Carduncellus, where diploids and tetraploids predominate, the only other reports of triploidy was for C. calvus and of hexaploidy, for C. caeruleus (L.) C. Presl. and C. pinnatus (Desf.) DC. (López González, 1990López González, G. 1990. Acerca de la clasificación natural del género Carthamus L., s.l. Anales del Jardín Botánico de Madrid 47: 11–34. ; Rice et al., 2015Rice, A., Glick, L., Abadi, S., Einhorn, M., Kopelman, N. M., Salman-Minkov, A., Mayzel, J., Chay, O. & Mayrose, I. 2015. The Chromosome Counts Database (CCDB) – a community resource of plant chromosome numbers. New Phytologist 206: 19–26. https://doi.org/10.1111/nph.13191. Retrieved October 2, 2020, from http://ccdb.tau.ac.il/). B chromosomes have been found in several species of Carduncellus and this could be one of the reasons for the wide variability in genome size found within the same ploidy level in phylogenetically closely related species, since dysploidy is not frequent in this genus (Vilatersana et al., 2000bVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x; Vilatersana, 2002Vilatersana, R. 2002. Delimitació genèrica del complex Carthamus-Carduncellus: un assaig de biosistemàtica i sistemàtica molecular. PhD Thesis, Universitat de Barcelona, Barcelona.). Our results suggest a loss of DNA per basic genome in polyploids (mean 1Cx = 1.66 pg) with respect to diploids (mean 1Cx = 1.94 pg), a phenomenon known as genome downsizing, largely observed in plants (Leitch & Bennett, 2004Leitch, I. J. & Bennett, M. D. 2004. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society 82: 651–663. https://doi.org/10.1111/j.1095-8312.2004.00349.x), present in the family Asteraceae (e.g. Pires et al., 2004Pires, J. C., Lim, K. Y., Kovařík, A., Matyásek, R., Boyd, A., Leitch, A. R., Leitch, I. J., Bennett, M. D., Soltis, P. S. & Soltis, D. E. 2004. Molecular cytogenetic analysis of recently evolved Tragopogon (Asteraceae) allopolyploids reveal a karyotype that is additive of the diploid progenitors. American Journal of Botany 91: 1022–1035. https://doi.org/10.3732/ajb.91.7.1022; Chrtek et al., 2009Chrtek, Jr. J., Zahradniček, J., Krak, K. & Fehrer, J. 2009. Genome size in Hieracium subgenus Hieracium (Asteraceae) is strongly correlated with major phylogenetic groups. Annals of Botany 104: 161–178. https://doi.org/10.1093/aob/mcp107; Pellicer et al., 2010Pellicer, J., Garcia, S., Canela, M. Á., Garnatje, T., Korobkov, A. A., Twibell, J. D. & Vallès, J. 2010. Genome size dynamics in Artemisia L. (Asteraceae): following the track of polyploidy. Plant Biology 12: 820–830. https://doi.org/10.1111/j.1438-8677.2009.00268.x; Vallès et al., 2013Vallès, J., Canela, M. Á., Garcia, S., Hidalgo, O., Pellicer, J., Sánchez-Jiménez, I., Siljak-Yakovlev, S., Vitales, D. & Garnatje, T. 2013. Genome size variation and evolution in the family Asteraceae. Caryologia 66: 221–235. https://doi.org/10.1080/00087114.2013.829690). The decrease of 1Cx values in the polyploid species has been found statistically significant by ANOVA test (F = 7.1914, p = 0.0143).
Genome size trends in the Carthamus-Carduncellus complex
The insular Femeniasia has a lower nuclear DNA content than Phonus, a continental sister genus with which it shares clade and chromosome number (Fig. 1, Table 2). The insularity may have played a role in the reduced genome size of a genus endemic of a Mediterranean island (Menorca) but further studies will be needed to confirm this hypothesis. Similar cases were reported in Carthamus (Garnatje et al., 2006Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050) and Cheirolophus (Garnatje et al., 2007Garnatje, T., Garcia, S. & Canela, M. Á. 2007. Genome size variation from a phylogenetic perspective in the genus Cheirolophus Cass. (Asteraceae): biogeographic implications. Plant Systematics and Evolution 264: 117–134. https://doi.org/10.1007/s00606-006-0489-7; Hidalgo et al., 2017Hidalgo, O., Vitales, D., Vallès, J., Garnatje, T., Siljak-Yakovlev, S., Leitch, I. J. & Pellicer J. 2017. Cytogenetic insights into an oceanic island radiation: The dramatic evolution of pre-existing traits in Cheirolophus (Asteraceae: Cardueae: Centaureinae). Taxon 66: 146–157. https://doi.org/10.12705/661.8) species, from the same tribe, as well as in other Asteraceae (Zahradníček et al., 2018Zahradníček, J., Chrtek, J., Ferreira, M. Z., Krahulcová, A. & Fehrer, J. 2018. Genome size variation in the genus Andryala (Hieraciinae, Asteraceae). Folia Geobotanica 53: 429–447. https://doi.org/10.1007/s12224-018-9330-7). This phenomenon has been attributed to island colonisation pressure (Suda et al., 2003Suda, J., Kyncl, T. & Freiova, R. 2003. Nuclear DNA amounts in Macaronesian angiosperms. Annals of Botany 92: 153–164. https://doi.org/10.1093/aob/mcg104) and to the higher facility of naturalisation of plants with smaller genomes (Kapralov & Filatov, 2011Kapralov, M. V. & Filatov, D. A. 2011. Does large genome size limit speciation in endemic island floras? Journal of Botany 2011: 458684. https://doi.org/10.1155/2011/458684).
Considering only the diploid taxa, the 1Cx average was 1.33 pg for Carthamus and 1.85 pg for Carduncellus-Femeniasia-Phonus clade. Statistically significant differences in the 1Cx values between Carthamus and Carduncellus (F = 99.8583, p = 0.0000) support the previously stated independent genomic evolution of these two lineages, although phylogenetic inferences have not yet fully resolved the relationships between Carthamus and the western group (Carduncellus, Femeniasia and Phonus; Vilatersana et al., 2000aVilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000a. Generic delimitation and phylogeny of the Carduncellus–Carthamus complex (Asteraceae) based on ITS sequences. Plant Systematics and Evolution 221: 89–105. https://doi.org/10.1007/BF01086383). For each ploidy level, genome size values in the Carduncellus-Femeniasia-Phonus clade are consistently higher than those of Carthamus; indeed, their range do not even overlap (Fig. 1B). This trend of higher genome sizes in perennial taxa compared to relatives with short life cycle is frequently observed in plants (for Asteraceae, see e.g. Hidalgo et al., 2008Hidalgo, O., Garcia-Jacas, N., Garnatje, T., Romashchenko, K., Susanna, A. & Siljak-Yakovlev, S. 2008. Extreme environmental conditions and phylogenetic inheritance: systematics of Myopordon and Oligochaeta (Asteraceae, Cardueae-Centaureinae). Taxon 57: 769–778. https://doi.org/10.1002/tax.573009; Siljak-Yakovlev et al., 2017Siljak-Yakovlev, S., Godelle, B., Zoldos, V., Vallès, J., Garnatje, T. & Hidalgo, O. 2017. Evolutionary implications of heterochromatin and rDNA in chromosome number and genome size changes during dysploidy: A case study in Reichardia genus. PLoS ONE 12: e0182318. https://doi.org/10.1371/journal.pone.0182318; Qiu et al., 2019Qiu, F., Baack, E. J., Whitney, K. D., Bock, D. G., Tetreault, H. M., Rieseberg, L. H. & Ungerer, M. C. 2019. Phylogenetic trends and environmental correlates of nuclear genome size variation in Helianthus sunflowers. New Phytologist 221: 1609–1618. https://doi.org/10.1111/nph.15465; but see Pellicer et al., 2014Pellicer, J., Hidalgo, O., Garnatje, T., Kondo, K. & Vallès, J. 2014. Life cycle versus systematic placement: phylogenetic and cytogenetic studies in annual Artemisia (Asteraceae, Anthemideae). Turkish Journal of Botany 38: 1112–1122. https://doi.org/10.3906/bot-1404-102). However, as suggested by Vitales et al. (2019Vitales, D., Fernández, P., Garnatje, T. & Garcia, S. 2019. Progress in the study of genome size evolution in Asteraceae: analysis of the last update. Database 2019: baz098. https://doi.org/10.1093/database/baz098), the observed associations between genome size and life cycle in Asteraceae could be better explained by phylogenetic relatedness between taxa. Yet, more data is needed, and also analysed in an evolutionary context, to establish such an association.
CONCLUSIONSTop
Genome size has proved to be a valuable tool for discriminating between closely related plant groups. The observed differences in the DNA amount between the two main clades of the Carthamus-Carduncellus complex, the Carthamus genus on the one hand and Carduncellus-Femeniasia-Phonus clade on the other, suggest that these genera have evolved independently. In this sense, our results give support to taxonomic treatments of the Carthamus-Carduncellus complex that would consider at least two genera.
ACKNOWLEDGEMENTSTop
We thank people mentioned in Table 2 for their help in plant collection, Chari González, Jaume Comas, Ricard Álvarez (Centres Científics i Tecnològics, Universitat de Barcelona) and Màrius Mumbrú (Laboratori de Botànica, Facultat de Farmàcia i Ciències de l’Alimentació, Universitat de Barcelona) for their assistance in flow cytometry measurements, Spencer C. Brown and Olivier Catrice (Institut des Sciences du Végétal, CNRS, Gif-surYvette) for supplying Petunia hybrida and Pisum sativum, used as internal standards, and Roi Rodríguez (Botanical Institute of Barcelona) for his technical support. This work has been supported by projects from the Spanish Government [(CGL2016-75694-P (AEI/FEDER, UE)] and the Generalitat de Catalunya (2017SGR1116). SG is the holder of a Ramón y Cajal contract (RYC-2014-16608).
REFERENCESTop
|
| 1. | Bancheva, S. & Greilhuber, J. 2006. Genome size in Bulgarian Centaurea s.l. (Asteraceae). Plant Systematics and Evolution 257: 95–117. https://doi.org/10.1007/s00606-005-0384-7 |
| 2. | Barres, L., Sanmartín, I., Anderson, C. L., Susanna, A., Buerki, S., Galbany-Casals, M. & Vilatersana, R. 2013. Reconstructing the evolution and biogeographic history of tribe Cardueae (Compositae). American Journal of Botany 100: 867–882. https://doi.org/10.3732/ajb.1200058 |
| 3. | Bureš, P., Wang, Y.-F., Horová, L. & Suda, J. 2004. Genome size variation in central European species of Cirsium (Compositae) and their natural hybrids. Annals of Botany 94: 353–363. https://doi.org/10.1093/aob/mch151 |
| 4. | Chrtek, Jr. J., Zahradniček, J., Krak, K. & Fehrer, J. 2009. Genome size in Hieracium subgenus Hieracium (Asteraceae) is strongly correlated with major phylogenetic groups. Annals of Botany 104: 161–178. https://doi.org/10.1093/aob/mcp107 |
| 5. | Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. https://doi.org/10.1038/nmeth.2109 |
| 6. | Doležel, J., Bartos, J., Voglmayr, H. & Greilhuber, J. 2003. Nuclear DNA content and genome size of trout and human. Cytometry 51A: 127–128. https://doi.org/10.1002/cyto.a.10013 |
| 7. | Doležel, J., Binarová, P. & Lucretti, S. 1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biologia Plantarum 3: 113–120. https://doi.org/10.1007/BF02907241 |
| 8. | Garcia, S., Canela M. Á., Garnatje, T., McArthur, E. D., Pellicer, J., Sanderson, S. C. & Vallès, J. 2008. Evolutionary and ecological implications of genome size in the North American endemic sagebrushes and allies (Artemisia, Asteraceae). Biological Journal of the Linnean Society 94: 631–649. https://doi.org/10.1111/j.1095-8312.2008.01001.x |
| 9. | Garcia, S., Garnatje, T., Twibell, J. D. & Vallès, J. 2006. Genome size variation in the Artemisia arborescens complex (Asteraceae, Anthemideae) and its cultivars. Genome 49: 244–253. https://doi.org/10.1139/g05-105 |
| 10. | Garnatje, T., Garcia, S. & Canela, M. Á. 2007. Genome size variation from a phylogenetic perspective in the genus Cheirolophus Cass. (Asteraceae): biogeographic implications. Plant Systematics and Evolution 264: 117–134. https://doi.org/10.1007/s00606-006-0489-7 |
| 11. | Garnatje, T., Garcia, S., Vilatersana, R. & Vallès, J. 2006. Genome size variation in the genus Carthamus (Asteraceae, Cardueae): Systematic implications and additive changes during allopolyploidization. Annals of Botany 97: 461–467. https://doi.org/10.1093/aob/mcj050 |
| 12. | Greuter, W. 2003. The Euro+Med treatment of Cardueae (Compositae) – generic concepts and required new names. Willdenowia 33: 49–61. https://doi.org/10.3372/wi.33.33104 |
| 13. | Hall, T. A. 1999. BioEdit, A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. |
| 14. | Hidalgo, O., Garcia-Jacas, N., Garnatje, T., Romashchenko, K., Susanna, A. & Siljak-Yakovlev, S. 2008. Extreme environmental conditions and phylogenetic inheritance: systematics of Myopordon and Oligochaeta (Asteraceae, Cardueae-Centaureinae). Taxon 57: 769–778. https://doi.org/10.1002/tax.573009 |
| 15. | Hidalgo, O., Vitales, D., Vallès, J., Garnatje, T., Siljak-Yakovlev, S., Leitch, I. J. & Pellicer J. 2017. Cytogenetic insights into an oceanic island radiation: The dramatic evolution of pre-existing traits in Cheirolophus (Asteraceae: Cardueae: Centaureinae). Taxon 66: 146–157. https://doi.org/10.12705/661.8 |
| 16. | Kapralov, M. V. & Filatov, D. A. 2011. Does large genome size limit speciation in endemic island floras? Journal of Botany 2011: 458684. https://doi.org/10.1155/2011/458684 |
| 17. | Leitch, I. J. & Bennett, M. D. 2004. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society 82: 651–663. https://doi.org/10.1111/j.1095-8312.2004.00349.x |
| 18. | López González, G. 1990. Acerca de la clasificación natural del género Carthamus L., s.l. Anales del Jardín Botánico de Madrid 47: 11–34. |
| 19. | López González, G. 2012. Sobre la clasificación del complejo Carthamus-Carduncellus (Asteraceae, Cardueae-Centaureinae) y su tratamiento en Flora iberica. Acta Botanica Malacitana 37: 79–92. https://doi.org/10.24310/abm.v37i0.2669 |
| 20. | Pegoraro, L., Baker, E. C., Aeschimann, D., Balant, M., Douzet, R., Garnatje, T., Guignard, M. S., Leitch, I. J., Leitch, A. R., Palazzesi, L., Theurillat, J. P., Hidalgo, O. & Pellicer, J. 2020. The correlation of phylogeny, elevation and ploidy on the incidence of apomixis in Asteraceae of the European Alps. Botanical Journal of the Linnean Society 194: 410–422. https://doi.org/10.1093/botlinnean/boaa058 |
| 21. | Pellicer, J., Garcia, S., Canela, M. Á., Garnatje, T., Korobkov, A. A., Twibell, J. D. & Vallès, J. 2010. Genome size dynamics in Artemisia L. (Asteraceae): following the track of polyploidy. Plant Biology 12: 820–830. https://doi.org/10.1111/j.1438-8677.2009.00268.x |
| 22. | Pellicer, J., Hidalgo, O., Garnatje, T., Kondo, K. & Vallès, J. 2014. Life cycle versus systematic placement: phylogenetic and cytogenetic studies in annual Artemisia (Asteraceae, Anthemideae). Turkish Journal of Botany 38: 1112–1122. https://doi.org/10.3906/bot-1404-102 |
| 23. | Pellicer, J. & Leitch, I. J. 2020. The Plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytologist 226: 301–305. https://doi.org/10.1111/nph.16261 |
| 24. | Pires, J. C., Lim, K. Y., Kovařík, A., Matyásek, R., Boyd, A., Leitch, A. R., Leitch, I. J., Bennett, M. D., Soltis, P. S. & Soltis, D. E. 2004. Molecular cytogenetic analysis of recently evolved Tragopogon (Asteraceae) allopolyploids reveal a karyotype that is additive of the diploid progenitors. American Journal of Botany 91: 1022–1035. https://doi.org/10.3732/ajb.91.7.1022 |
| 25. | Qiu, F., Baack, E. J., Whitney, K. D., Bock, D. G., Tetreault, H. M., Rieseberg, L. H. & Ungerer, M. C. 2019. Phylogenetic trends and environmental correlates of nuclear genome size variation in Helianthus sunflowers. New Phytologist 221: 1609–1618. https://doi.org/10.1111/nph.15465 |
| 26. | R Core Team 2016. R: a language and environment for statistical computing. Foundation for Statistical Computing, Vienna. Version 3.6.2. Retrieved December 12, 2019, from http://www.R-project.org |
| 27. | Revell, L. J. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. https://doi.org/10.1111/j.2041-210X.2011.00169.x |
| 28. | Rice, A., Glick, L., Abadi, S., Einhorn, M., Kopelman, N. M., Salman-Minkov, A., Mayzel, J., Chay, O. & Mayrose, I. 2015. The Chromosome Counts Database (CCDB) – a community resource of plant chromosome numbers. New Phytologist 206: 19–26. https://doi.org/10.1111/nph.13191. Retrieved October 2, 2020, from http://ccdb.tau.ac.il/ |
| 29. | Ronquist, F., Teslenko, M., Mark, P., Ayres, D., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A. & Huelsenbeck, J. P. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029 |
| 30. | Siljak-Yakovlev, S., Godelle, B., Zoldos, V., Vallès, J., Garnatje, T. & Hidalgo, O. 2017. Evolutionary implications of heterochromatin and rDNA in chromosome number and genome size changes during dysploidy: A case study in Reichardia genus. PLoS ONE 12: e0182318. https://doi.org/10.1371/journal.pone.0182318 |
| 31. | Suda, J., Krahulcová, A., Trávnícek, P., Rosenbaumová, R., Peckert, T. & Krahulec, F. 2007. Genome size variation and species relationships in Hieracium sub-genus Pilosella (Asteraceae) as inferred by flow cytometry. Annals of Botany 100: 1323–1335. https://doi.org/10.1093/aob/mcm218 |
| 32. | Suda, J., Kyncl, T. & Freiova, R. 2003. Nuclear DNA amounts in Macaronesian angiosperms. Annals of Botany 92: 153–164. https://doi.org/10.1093/aob/mcg104 |
| 33. | Torrell, M. & Vallès, J. 2001. Genome size in 21 Artemisia L. species (Asteraceae, Anthemideae): Systematic, evolutionary, and ecological implications. Genome 44: 231–238. https://doi.org/10.1139/g01-004 |
| 34. | Trávníček, P., Kirschner, J., Chudáčková, H., Rooks, F. & Štěpánek, J. 2013. Substantial genome size variation in Taraxacum stenocephalum (Asteraceae, Lactuceae). Folia Geobotanica 48: 271–284. https://doi.org/10.1007/s12224-013-9151-7 |
| 35. | Vallès, J., Canela, M. Á., Garcia, S., Hidalgo, O., Pellicer, J., Sánchez-Jiménez, I., Siljak-Yakovlev, S., Vitales, D. & Garnatje, T. 2013. Genome size variation and evolution in the family Asteraceae. Caryologia 66: 221–235. https://doi.org/10.1080/00087114.2013.829690 |
| 36. | Vilatersana, R. 2002. Delimitació genèrica del complex Carthamus-Carduncellus: un assaig de biosistemàtica i sistemàtica molecular. PhD Thesis, Universitat de Barcelona, Barcelona. |
| 37. | Vilatersana, R., Brysting, A. K. & Brochmann, C. 2007. Molecular evidence for hybrid origins of the invasive polyploids Carthamus creticus and C. turkestanicus (Cardueae, Asteraceae). Molecular Phylogenetics and Evolution 44: 610–621. https://doi.org/10.1016/j.ympev.2007.05.008 |
| 38. | Vilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000a. Generic delimitation and phylogeny of the Carduncellus–Carthamus complex (Asteraceae) based on ITS sequences. Plant Systematics and Evolution 221: 89–105. https://doi.org/10.1007/BF01086383 |
| 39. | Vilatersana, R., Susanna, A., Garcia-Jacas, N. & Garnatje, T. 2000b. Karyology, generic delineation and dysploidy in the genera Carduncellus, Carthamus and Phonus (Asteraceae). Botanical Journal of the Linnean Society 134: 425–438. https://doi.org/10.1111/j.1095-8339.2000.tb00539.x |
| 40. | Vitales, D., Álvarez, I., Garcia, S., Hidalgo, O., Nieto Feliner, G., Pellicer, J., Vallès, J. & Garnatje, T. 2020. Genome size variation at constant chromosome number is not correlated with repetitive DNA dynamism in Anacyclus (Asteraceae). Annals of Botany 125: 611–623. https://doi.org/10.1093/aob/mcz183 |
| 41. | Vitales, D., Fernández, P., Garnatje, T. & Garcia, S. 2019. Progress in the study of genome size evolution in Asteraceae: analysis of the last update. Database 2019: baz098. https://doi.org/10.1093/database/baz098 |
| 42. | Wickham, H. 2016. ggplot2: elegant graphics for data analysis. Springer, New York. https://doi.org/10.1007/978-3-319-24277-4 |
| 43. | Zahradníček, J., Chrtek, J., Ferreira, M. Z., Krahulcová, A. & Fehrer, J. 2018. Genome size variation in the genus Andryala (Hieraciinae, Asteraceae). Folia Geobotanica 53: 429–447. https://doi.org/10.1007/s12224-018-9330-7 |